ISLR习题:线性回归 - Boston数据集
目录
本文源自《统计学习导论:基于R语言应用》(ISLR) 第三章习题
Boston 数据集
library(MASS)
attach(Boston)
head(Boston)
crim zn indus chas nox rm age dis rad tax ptratio black lstat medv
1 0.00632 18 2.31 0 0.538 6.575 65.2 4.0900 1 296 15.3 396.90 4.98 24.0
2 0.02731 0 7.07 0 0.469 6.421 78.9 4.9671 2 242 17.8 396.90 9.14 21.6
3 0.02729 0 7.07 0 0.469 7.185 61.1 4.9671 2 242 17.8 392.83 4.03 34.7
4 0.03237 0 2.18 0 0.458 6.998 45.8 6.0622 3 222 18.7 394.63 2.94 33.4
5 0.06905 0 2.18 0 0.458 7.147 54.2 6.0622 3 222 18.7 396.90 5.33 36.2
6 0.02985 0 2.18 0 0.458 6.430 58.7 6.0622 3 222 18.7 394.12 5.21 28.7
简单线性回归
zn
lm_fit_zn <- lm(crim ~ zn)
summary(lm_fit_zn)
Call:
lm(formula = crim ~ zn)
Residuals:
Min 1Q Median 3Q Max
-4.429 -4.222 -2.620 1.250 84.523
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 4.45369 0.41722 10.675 < 2e-16 ***
zn -0.07393 0.01609 -4.594 5.51e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 8.435 on 504 degrees of freedom
Multiple R-squared: 0.04019, Adjusted R-squared: 0.03828
F-statistic: 21.1 on 1 and 504 DF, p-value: 5.506e-06
indus
lm_fit_indus <- lm(crim ~ indus)
summary(lm_fit_indus)
Call:
lm(formula = crim ~ indus)
Residuals:
Min 1Q Median 3Q Max
-11.972 -2.698 -0.736 0.712 81.813
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.06374 0.66723 -3.093 0.00209 **
indus 0.50978 0.05102 9.991 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.866 on 504 degrees of freedom
Multiple R-squared: 0.1653, Adjusted R-squared: 0.1637
F-statistic: 99.82 on 1 and 504 DF, p-value: < 2.2e-16
chas
lm_fit_chas <- lm(crim ~ chas)
summary(lm_fit_chas)
Call:
lm(formula = crim ~ chas)
Residuals:
Min 1Q Median 3Q Max
-3.738 -3.661 -3.435 0.018 85.232
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.7444 0.3961 9.453 <2e-16 ***
chas -1.8928 1.5061 -1.257 0.209
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 8.597 on 504 degrees of freedom
Multiple R-squared: 0.003124, Adjusted R-squared: 0.001146
F-statistic: 1.579 on 1 and 504 DF, p-value: 0.2094
没有显著性
nox
lm_fit_nox <- lm(crim ~ nox)
summary(lm_fit_nox)
Call:
lm(formula = crim ~ nox)
Residuals:
Min 1Q Median 3Q Max
-12.371 -2.738 -0.974 0.559 81.728
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -13.720 1.699 -8.073 5.08e-15 ***
nox 31.249 2.999 10.419 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.81 on 504 degrees of freedom
Multiple R-squared: 0.1772, Adjusted R-squared: 0.1756
F-statistic: 108.6 on 1 and 504 DF, p-value: < 2.2e-16
rm
lm_fit_rm <- lm(crim ~ rm)
summary(lm_fit_rm)
Call:
lm(formula = crim ~ rm)
Residuals:
Min 1Q Median 3Q Max
-6.604 -3.952 -2.654 0.989 87.197
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 20.482 3.365 6.088 2.27e-09 ***
rm -2.684 0.532 -5.045 6.35e-07 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 8.401 on 504 degrees of freedom
Multiple R-squared: 0.04807, Adjusted R-squared: 0.04618
F-statistic: 25.45 on 1 and 504 DF, p-value: 6.347e-07
age
lm_fit_age <- lm(crim ~ age)
summary(lm_fit_age)
Call:
lm(formula = crim ~ age)
Residuals:
Min 1Q Median 3Q Max
-6.789 -4.257 -1.230 1.527 82.849
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3.77791 0.94398 -4.002 7.22e-05 ***
age 0.10779 0.01274 8.463 2.85e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 8.057 on 504 degrees of freedom
Multiple R-squared: 0.1244, Adjusted R-squared: 0.1227
F-statistic: 71.62 on 1 and 504 DF, p-value: 2.855e-16
dis
lm_fit_dis <- lm(crim ~ dis)
summary(lm_fit_dis)
Call:
lm(formula = crim ~ dis)
Residuals:
Min 1Q Median 3Q Max
-6.708 -4.134 -1.527 1.516 81.674
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 9.4993 0.7304 13.006 <2e-16 ***
dis -1.5509 0.1683 -9.213 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.965 on 504 degrees of freedom
Multiple R-squared: 0.1441, Adjusted R-squared: 0.1425
F-statistic: 84.89 on 1 and 504 DF, p-value: < 2.2e-16
rad
lm_fit_rad <- lm(crim ~ rad)
summary(lm_fit_rad)
Call:
lm(formula = crim ~ rad)
Residuals:
Min 1Q Median 3Q Max
-10.164 -1.381 -0.141 0.660 76.433
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.28716 0.44348 -5.157 3.61e-07 ***
rad 0.61791 0.03433 17.998 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 6.718 on 504 degrees of freedom
Multiple R-squared: 0.3913, Adjusted R-squared: 0.39
F-statistic: 323.9 on 1 and 504 DF, p-value: < 2.2e-16
tax
lm_fit_tax <- lm(crim ~ tax)
summary(lm_fit_tax)
Call:
lm(formula = crim ~ tax)
Residuals:
Min 1Q Median 3Q Max
-12.513 -2.738 -0.194 1.065 77.696
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -8.528369 0.815809 -10.45 <2e-16 ***
tax 0.029742 0.001847 16.10 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 6.997 on 504 degrees of freedom
Multiple R-squared: 0.3396, Adjusted R-squared: 0.3383
F-statistic: 259.2 on 1 and 504 DF, p-value: < 2.2e-16
ptratio
lm_fit_ptratio <- lm(crim ~ ptratio)
summary(lm_fit_ptratio)
Call:
lm(formula = crim ~ ptratio)
Residuals:
Min 1Q Median 3Q Max
-7.654 -3.985 -1.912 1.825 83.353
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -17.6469 3.1473 -5.607 3.40e-08 ***
ptratio 1.1520 0.1694 6.801 2.94e-11 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 8.24 on 504 degrees of freedom
Multiple R-squared: 0.08407, Adjusted R-squared: 0.08225
F-statistic: 46.26 on 1 and 504 DF, p-value: 2.943e-11
black
lm_fit_black <- lm(crim ~ black)
summary(lm_fit_black)
Call:
lm(formula = crim ~ black)
Residuals:
Min 1Q Median 3Q Max
-13.756 -2.299 -2.095 -1.296 86.822
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 16.553529 1.425903 11.609 <2e-16 ***
black -0.036280 0.003873 -9.367 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.946 on 504 degrees of freedom
Multiple R-squared: 0.1483, Adjusted R-squared: 0.1466
F-statistic: 87.74 on 1 and 504 DF, p-value: < 2.2e-16
lstat
lm_fit_lstat <- lm(crim ~ lstat)
summary(lm_fit_lstat)
Call:
lm(formula = crim ~ lstat)
Residuals:
Min 1Q Median 3Q Max
-13.925 -2.822 -0.664 1.079 82.862
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3.33054 0.69376 -4.801 2.09e-06 ***
lstat 0.54880 0.04776 11.491 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.664 on 504 degrees of freedom
Multiple R-squared: 0.2076, Adjusted R-squared: 0.206
F-statistic: 132 on 1 and 504 DF, p-value: < 2.2e-16
medv
lm_fit_medv <- lm(crim ~ medv)
summary(lm_fit_medv)
Call:
lm(formula = crim ~ medv)
Residuals:
Min 1Q Median 3Q Max
-9.071 -4.022 -2.343 1.298 80.957
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 11.79654 0.93419 12.63 <2e-16 ***
medv -0.36316 0.03839 -9.46 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.934 on 504 degrees of freedom
Multiple R-squared: 0.1508, Adjusted R-squared: 0.1491
F-statistic: 89.49 on 1 and 504 DF, p-value: < 2.2e-16
多元线性回归
lm_fit_multi <- lm(crim~., data=Boston)
summary(lm_fit_multi)
Call:
lm(formula = crim ~ ., data = Boston)
Residuals:
Min 1Q Median 3Q Max
-9.924 -2.120 -0.353 1.019 75.051
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 17.033228 7.234903 2.354 0.018949 *
zn 0.044855 0.018734 2.394 0.017025 *
indus -0.063855 0.083407 -0.766 0.444294
chas -0.749134 1.180147 -0.635 0.525867
nox -10.313535 5.275536 -1.955 0.051152 .
rm 0.430131 0.612830 0.702 0.483089
age 0.001452 0.017925 0.081 0.935488
dis -0.987176 0.281817 -3.503 0.000502 ***
rad 0.588209 0.088049 6.680 6.46e-11 ***
tax -0.003780 0.005156 -0.733 0.463793
ptratio -0.271081 0.186450 -1.454 0.146611
black -0.007538 0.003673 -2.052 0.040702 *
lstat 0.126211 0.075725 1.667 0.096208 .
medv -0.198887 0.060516 -3.287 0.001087 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 6.439 on 492 degrees of freedom
Multiple R-squared: 0.454, Adjusted R-squared: 0.4396
F-statistic: 31.47 on 13 and 492 DF, p-value: < 2.2e-16
具有显著性的变量:
- zn
- dis
- rad
- black
- medv
对比系数
col_names <- colnames(Boston)[-1]
get_coef <- function(name) {
coefficients(get(paste("lm_fit_", name, sep="")))[2]
}
simple_coefs <- sapply(col_names, get_coef)
simple_coefs
zn.zn indus.indus chas.chas nox.nox rm.rm
-0.07393498 0.50977633 -1.89277655 31.24853120 -2.68405122
age.age dis.dis rad.rad tax.tax ptratio.ptratio
0.10778623 -1.55090168 0.61791093 0.02974225 1.15198279
black.black lstat.lstat medv.medv
-0.03627964 0.54880478 -0.36315992
multi_coefs <- coefficients(lm_fit_multi)[-1]
multi_coefs
zn indus chas nox rm
0.044855215 -0.063854824 -0.749133611 -10.313534912 0.430130506
age dis rad tax ptratio
0.001451643 -0.987175726 0.588208591 -0.003780016 -0.271080558
black lstat medv
-0.007537505 0.126211376 -0.198886821
plot(simple_coefs, multi_coefs)
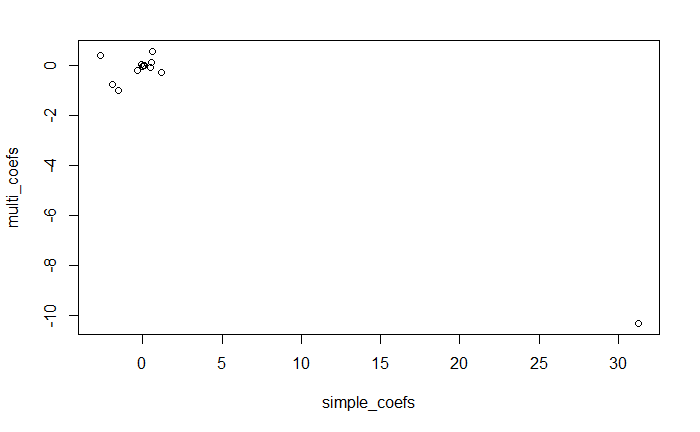
对 nox 的估计系数相差很大
非线性
zn
lm_fit_poly_zn <- lm(crim ~ poly(zn, 3))
summary(lm_fit_poly_zn)
Call:
lm(formula = crim ~ poly(zn, 3))
Residuals:
Min 1Q Median 3Q Max
-4.821 -4.614 -1.294 0.473 84.130
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.6135 0.3722 9.709 < 2e-16 ***
poly(zn, 3)1 -38.7498 8.3722 -4.628 4.7e-06 ***
poly(zn, 3)2 23.9398 8.3722 2.859 0.00442 **
poly(zn, 3)3 -10.0719 8.3722 -1.203 0.22954
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 8.372 on 502 degrees of freedom
Multiple R-squared: 0.05824, Adjusted R-squared: 0.05261
F-statistic: 10.35 on 3 and 502 DF, p-value: 1.281e-06
indus
lm_fit_poly_indus <- lm(crim ~ poly(indus, 3))
summary(lm_fit_poly_indus)
Call:
lm(formula = crim ~ poly(indus, 3))
Residuals:
Min 1Q Median 3Q Max
-8.278 -2.514 0.054 0.764 79.713
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.614 0.330 10.950 < 2e-16 ***
poly(indus, 3)1 78.591 7.423 10.587 < 2e-16 ***
poly(indus, 3)2 -24.395 7.423 -3.286 0.00109 **
poly(indus, 3)3 -54.130 7.423 -7.292 1.2e-12 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.423 on 502 degrees of freedom
Multiple R-squared: 0.2597, Adjusted R-squared: 0.2552
F-statistic: 58.69 on 3 and 502 DF, p-value: < 2.2e-16
chas
lm_fit_poly_chas <- lm(crim ~ poly(chas, 3))
summary(lm_fit_poly_chas)
无法执行
nox
lm_fit_poly_nox <- lm(crim ~ poly(nox, 3))
summary(lm_fit_poly_nox)
Call:
lm(formula = crim ~ poly(nox, 3))
Residuals:
Min 1Q Median 3Q Max
-9.110 -2.068 -0.255 0.739 78.302
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.6135 0.3216 11.237 < 2e-16 ***
poly(nox, 3)1 81.3720 7.2336 11.249 < 2e-16 ***
poly(nox, 3)2 -28.8286 7.2336 -3.985 7.74e-05 ***
poly(nox, 3)3 -60.3619 7.2336 -8.345 6.96e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.234 on 502 degrees of freedom
Multiple R-squared: 0.297, Adjusted R-squared: 0.2928
F-statistic: 70.69 on 3 and 502 DF, p-value: < 2.2e-16
rm
lm_fit_poly_rm <- lm(crim ~ poly(rm, 3))
summary(lm_fit_poly_rm)
Call:
lm(formula = crim ~ poly(rm, 3))
Residuals:
Min 1Q Median 3Q Max
-18.485 -3.468 -2.221 -0.015 87.219
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.6135 0.3703 9.758 < 2e-16 ***
poly(rm, 3)1 -42.3794 8.3297 -5.088 5.13e-07 ***
poly(rm, 3)2 26.5768 8.3297 3.191 0.00151 **
poly(rm, 3)3 -5.5103 8.3297 -0.662 0.50858
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 8.33 on 502 degrees of freedom
Multiple R-squared: 0.06779, Adjusted R-squared: 0.06222
F-statistic: 12.17 on 3 and 502 DF, p-value: 1.067e-07
age
lm_fit_poly_age <- lm(crim ~ poly(age, 3))
summary(lm_fit_poly_age)
Call:
lm(formula = crim ~ poly(age, 3))
Residuals:
Min 1Q Median 3Q Max
-9.762 -2.673 -0.516 0.019 82.842
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.6135 0.3485 10.368 < 2e-16 ***
poly(age, 3)1 68.1820 7.8397 8.697 < 2e-16 ***
poly(age, 3)2 37.4845 7.8397 4.781 2.29e-06 ***
poly(age, 3)3 21.3532 7.8397 2.724 0.00668 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.84 on 502 degrees of freedom
Multiple R-squared: 0.1742, Adjusted R-squared: 0.1693
F-statistic: 35.31 on 3 and 502 DF, p-value: < 2.2e-16
dis
lm_fit_poly_dis <- lm(crim ~ poly(dis, 3))
summary(lm_fit_poly_dis)
Call:
lm(formula = crim ~ poly(dis, 3))
Residuals:
Min 1Q Median 3Q Max
-10.757 -2.588 0.031 1.267 76.378
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.6135 0.3259 11.087 < 2e-16 ***
poly(dis, 3)1 -73.3886 7.3315 -10.010 < 2e-16 ***
poly(dis, 3)2 56.3730 7.3315 7.689 7.87e-14 ***
poly(dis, 3)3 -42.6219 7.3315 -5.814 1.09e-08 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.331 on 502 degrees of freedom
Multiple R-squared: 0.2778, Adjusted R-squared: 0.2735
F-statistic: 64.37 on 3 and 502 DF, p-value: < 2.2e-16
rad
lm_fit_poly_rad <- lm(crim ~ poly(rad, 3))
summary(lm_fit_poly_rad)
Call:
lm(formula = crim ~ poly(rad, 3))
Residuals:
Min 1Q Median 3Q Max
-10.381 -0.412 -0.269 0.179 76.217
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.6135 0.2971 12.164 < 2e-16 ***
poly(rad, 3)1 120.9074 6.6824 18.093 < 2e-16 ***
poly(rad, 3)2 17.4923 6.6824 2.618 0.00912 **
poly(rad, 3)3 4.6985 6.6824 0.703 0.48231
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 6.682 on 502 degrees of freedom
Multiple R-squared: 0.4, Adjusted R-squared: 0.3965
F-statistic: 111.6 on 3 and 502 DF, p-value: < 2.2e-16
tax
lm_fit_poly_tax <- lm(crim ~ poly(tax, 3))
summary(lm_fit_poly_tax)
Call:
lm(formula = crim ~ poly(tax, 3))
Residuals:
Min 1Q Median 3Q Max
-13.273 -1.389 0.046 0.536 76.950
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.6135 0.3047 11.860 < 2e-16 ***
poly(tax, 3)1 112.6458 6.8537 16.436 < 2e-16 ***
poly(tax, 3)2 32.0873 6.8537 4.682 3.67e-06 ***
poly(tax, 3)3 -7.9968 6.8537 -1.167 0.244
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 6.854 on 502 degrees of freedom
Multiple R-squared: 0.3689, Adjusted R-squared: 0.3651
F-statistic: 97.8 on 3 and 502 DF, p-value: < 2.2e-16
ptratio
lm_fit_poly_ptratio <- lm(crim ~ poly(ptratio, 3))
summary(lm_fit_poly_ptratio)
Call:
lm(formula = crim ~ poly(ptratio, 3))
Residuals:
Min 1Q Median 3Q Max
-6.833 -4.146 -1.655 1.408 82.697
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.614 0.361 10.008 < 2e-16 ***
poly(ptratio, 3)1 56.045 8.122 6.901 1.57e-11 ***
poly(ptratio, 3)2 24.775 8.122 3.050 0.00241 **
poly(ptratio, 3)3 -22.280 8.122 -2.743 0.00630 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 8.122 on 502 degrees of freedom
Multiple R-squared: 0.1138, Adjusted R-squared: 0.1085
F-statistic: 21.48 on 3 and 502 DF, p-value: 4.171e-13
black
lm_fit_poly_black <- lm(crim ~ poly(black, 3))
summary(lm_fit_poly_black)
Call:
lm(formula = crim ~ poly(black, 3))
Residuals:
Min 1Q Median 3Q Max
-13.096 -2.343 -2.128 -1.439 86.790
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.6135 0.3536 10.218 <2e-16 ***
poly(black, 3)1 -74.4312 7.9546 -9.357 <2e-16 ***
poly(black, 3)2 5.9264 7.9546 0.745 0.457
poly(black, 3)3 -4.8346 7.9546 -0.608 0.544
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.955 on 502 degrees of freedom
Multiple R-squared: 0.1498, Adjusted R-squared: 0.1448
F-statistic: 29.49 on 3 and 502 DF, p-value: < 2.2e-16
lstat
lm_fit_poly_lstat <- lm(crim ~ poly(lstat, 3))
summary(lm_fit_poly_lstat)
Call:
lm(formula = crim ~ poly(lstat, 3))
Residuals:
Min 1Q Median 3Q Max
-15.234 -2.151 -0.486 0.066 83.353
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.6135 0.3392 10.654 <2e-16 ***
poly(lstat, 3)1 88.0697 7.6294 11.543 <2e-16 ***
poly(lstat, 3)2 15.8882 7.6294 2.082 0.0378 *
poly(lstat, 3)3 -11.5740 7.6294 -1.517 0.1299
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.629 on 502 degrees of freedom
Multiple R-squared: 0.2179, Adjusted R-squared: 0.2133
F-statistic: 46.63 on 3 and 502 DF, p-value: < 2.2e-16
medv
lm_fit_poly_medv <- lm(crim ~ poly(medv, 3))
summary(lm_fit_poly_medv)
Call:
lm(formula = crim ~ poly(medv, 3))
Residuals:
Min 1Q Median 3Q Max
-24.427 -1.976 -0.437 0.439 73.655
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 3.614 0.292 12.374 < 2e-16 ***
poly(medv, 3)1 -75.058 6.569 -11.426 < 2e-16 ***
poly(medv, 3)2 88.086 6.569 13.409 < 2e-16 ***
poly(medv, 3)3 -48.033 6.569 -7.312 1.05e-12 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 6.569 on 502 degrees of freedom
Multiple R-squared: 0.4202, Adjusted R-squared: 0.4167
F-statistic: 121.3 on 3 and 502 DF, p-value: < 2.2e-16
不只一个变量有非线性关系
参考
https://github.com/perillaroc/islr-study
ISLR实验系列文章
线性回归
分类
重抽样方法
线性模型选择与正则化